Is a barrier-free, open-source, ongoing phage and strain collection available for your biological question of interest.
It is also expandable. Your Klebsiella-targeting phages and Klebsiella strains are welcome to form part of the collection.
Its contents
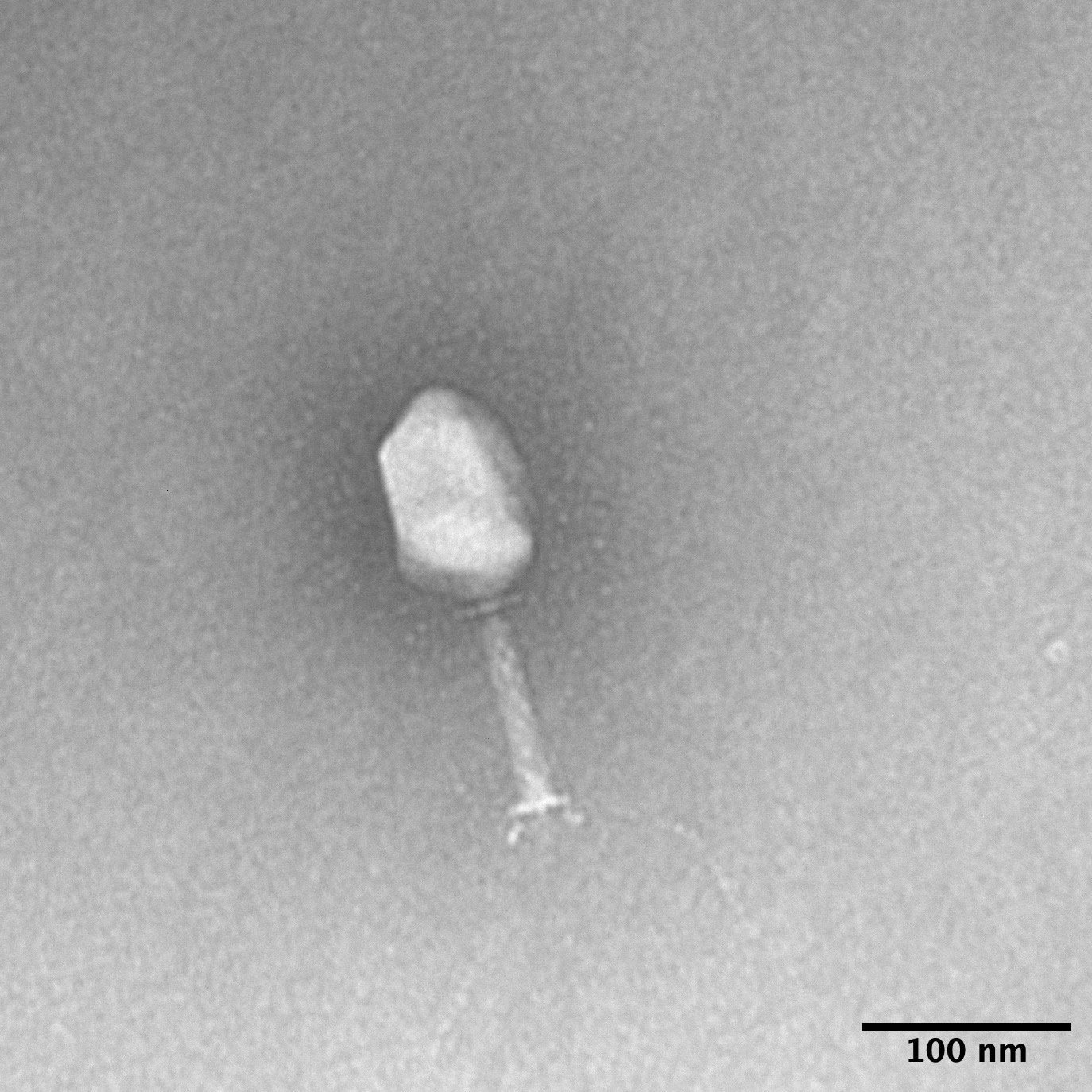
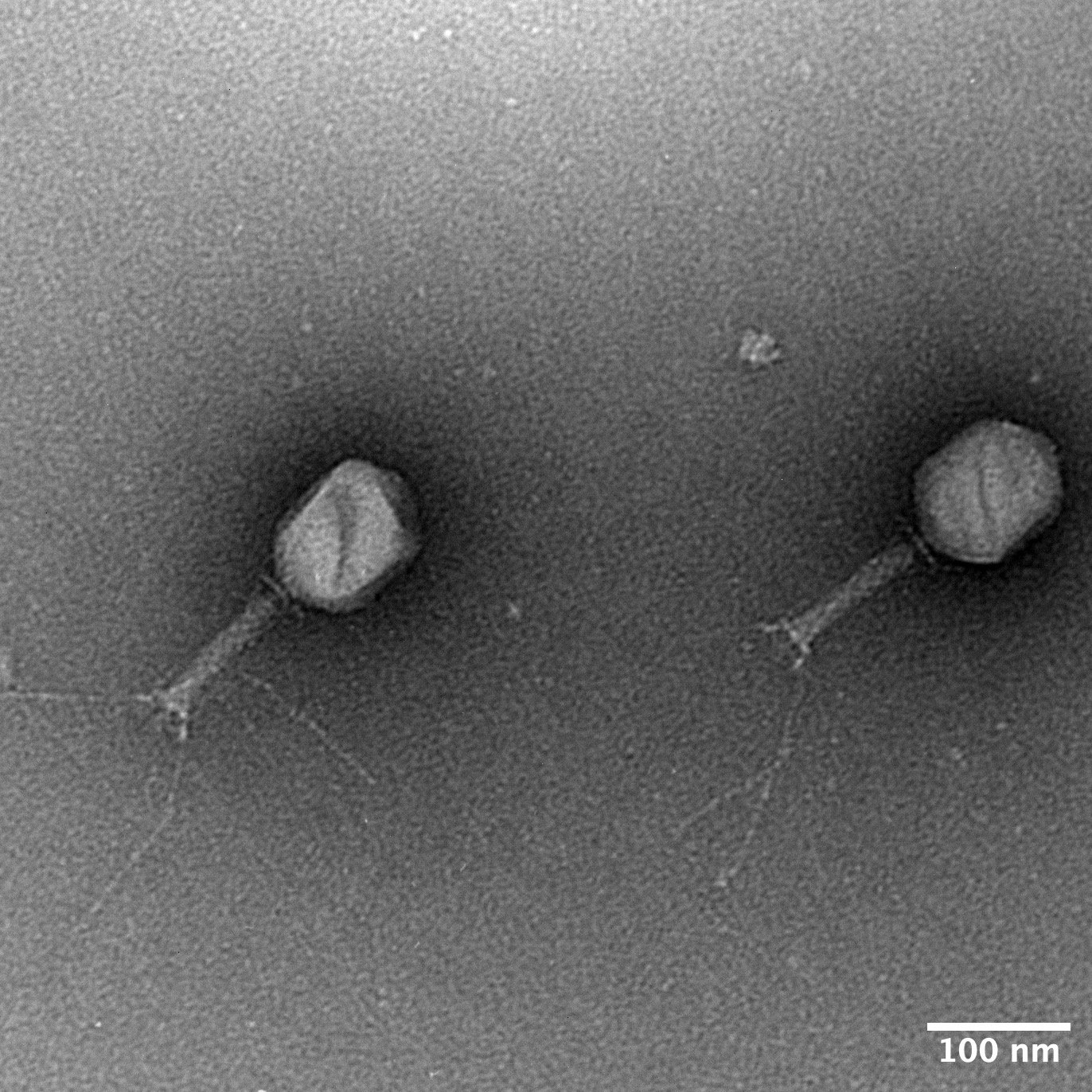
KlebPhaCol currently consists of 53 Klebsiella-targeting phages & 74 Klebsiella clinical strains along with their associated data, including:
Genomic sequences
Annotation files
Protein sequences
Complete characterization (incl. Host range, receptor data, genomic typing for strains, taxonomic classification)
Comparative analyses
TEM images and measurements
Your research output! All research contributions will be included too.
Click here to see how you can contribute.
Our Vision
KlebPhaCol was born out of enthusiasm for open science and community-based data contribution.
Our lab is interested in the capsulated Gram-negative pathogen and pathobiont Klebsiella pneumoniae due to its highly relevant clinical space, and complex biology. It is commonly known for its involvement in nosocomial and community-acquired infections. These infections can range from pneumonia, urinary tract, and soft tissue infections to more severe conditions such as septicemia, liver abscess, and sepsis, particularly for the hypervirulent strains. K. pneumoniae is also recognized as one of the six “ESKAPE” pathogens in need of new antimicrobials. Recently, gut microbial colonization by this bacteria has been linked to inflammatory diseases like inflammatory bowel diseases and primary sclerosing cholangitis. As a result, bacterio(phages) have become increasingly relevant as potent therapeutics for treating K. pneumoniae infections and gut colonization. However, the high variability of the Klebsiella capsule, with >79 known capsular (K) types to date, poses a significant challenge for receptor-mediated strategies targeting this pathogen (e.g. vaccine developments and phage therapy). Thus, it is crucial to understand phage-mediated interactions to ensure successful therapeutic applications. Unfortunately, this understanding is currently limited, partly due to the lack of a representative and accessible collection of Klebsiella phages and strains.
This is why we sought to isolate new phages against this pathogen and pathobiont and make them freely available, along with the clinical strains, for continued research outputs within the community*.
We envision this collection to be widely used to deepen our understanding of phage-bacteria-human interactions in the context of encapsulated bacteria, for which Klebsiella is a great model. We also hope the community is encouraged to offer previous and new Klebsiella phages and strains to KlebPhaCol to increase the research potential and scope. To maintain the open science movement, all contributors will get to share their data on this platform with recognition (via a DOI). See more on how to contribute here.
We are interested in making this a sustainable ongoing resource and as such, we are committed to a 5-year revision period of the collection to publish added contributions in a peer-reviewed journal.
*See our Terms of Use here.
We need you.
If our vision resonates with you, please do not hesitate to contact us with any further questions.
About Us
KlebPhaCol is a project led by the Nobrega Lab. We are based at the University of Southampton in the UK.
The progression of the field of Phage therapy is crucial during this threatening era of escalating antimicrobial resistance. We are deeply enthusiastic about the open science movement we are currently witnessing, and we believe a community-based open-source platform is the way to move Phage Therapy forward and reach important life-saving milestones. It is with this in mind that we created KlebPhaCol. We hope this inspires you to join the community, contribute, and help move Phage Therapy forward.
Our Sponsors
This research was primarily funded by Bowel Research UK, Wessex Medical Research, and SoCoBio DTP.